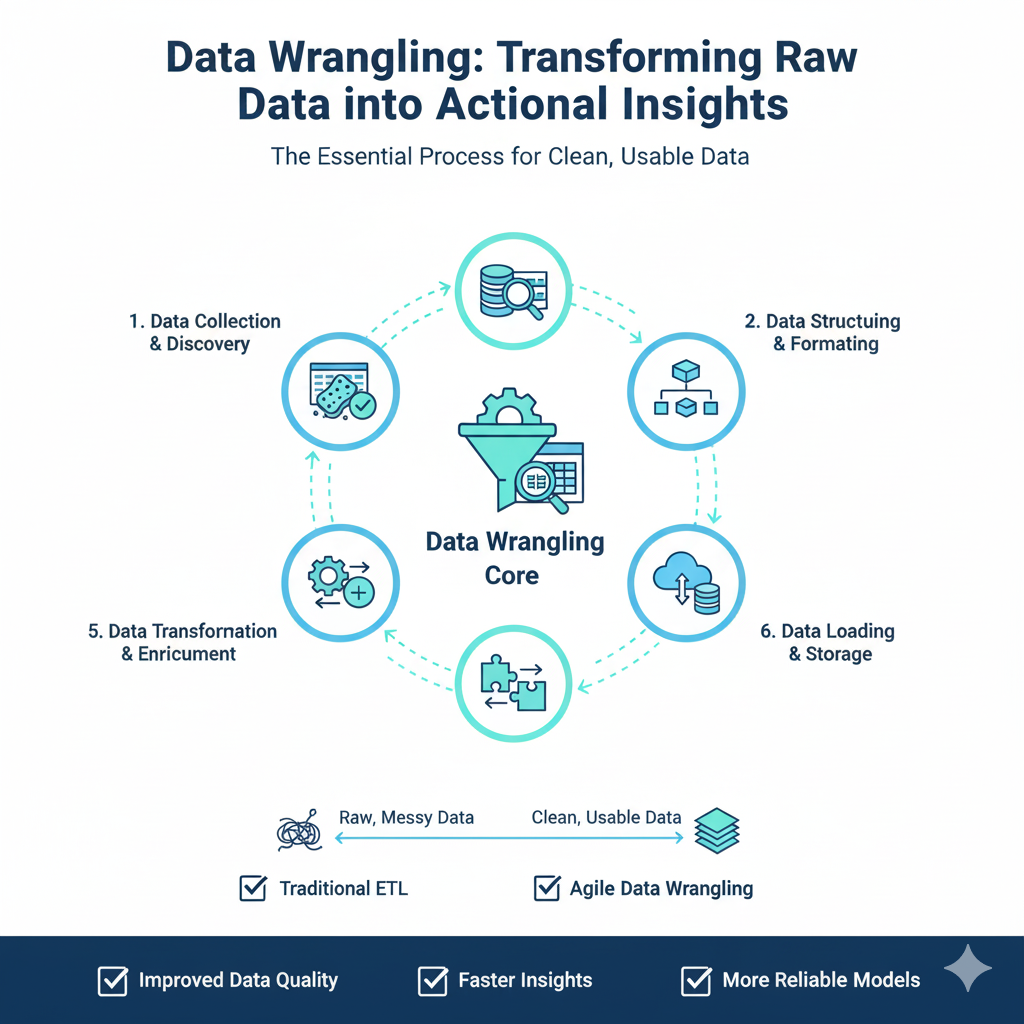
Introduction: The Critical Foundation of Data Wrangling
In the modern data-driven landscape, Data wrangling has emerged as the most crucial and time-consuming phase of any data analysis or machine learning project. Often consuming up to 80% of a data scientist’s time, Data wrangling represents the essential process of cleaning, structuring, and enriching raw data into a format suitable for analysis and modeling. The quality of Data wrangling directly determines the success of subsequent analytical endeavors—garbage in, garbage out remains as true today as it was in the earliest days of computing.
Data wrangling encompasses a comprehensive set of techniques and processes that transform disorganized, incomplete, and often messy real-world data into clean, structured datasets ready for analysis. This process involves handling missing values, correcting data types, dealing with outliers, merging datasets, creating new features, and ensuring data consistency. The sophistication of Data wrangling has grown exponentially as data sources have multiplied in volume, variety, and velocity. Today’s data professionals must wrangle everything from structured CSV files and database extracts to unstructured text, JSON APIs, and real-time streaming data.
The importance of mastering Data wrangling cannot be overstated. According to recent surveys from leading data science platforms, data scientists spend approximately 60-80% of their time on Data wrangling tasks. This time investment pays enormous dividends: properly wrangled data leads to more accurate models, more reliable insights, and more trustworthy business decisions. Conversely, poor Data wrangling can introduce biases, hide important patterns, and lead to completely erroneous conclusions.
This comprehensive guide will walk you through ten essential steps for effective Data wrangling using both Python and R, the two dominant programming languages in the data science ecosystem. We’ll explore each step in depth, providing practical code examples, real-world scenarios, and expert tips that you can immediately apply to your own data projects. Whether you’re working with small datasets on your local machine or big data in distributed computing environments, these Data wrangling principles will form the foundation of your data analysis success.
Step 1: Comprehensive Data Assessment and Understanding

The Foundation of Effective Data Wrangling
Before any transformation occurs, thorough data assessment sets the stage for successful Data wrangling. This critical first step involves understanding your data’s structure, quality, and characteristics. Comprehensive assessment prevents wasted effort and ensures that your Data wrangling approach addresses the actual issues present in your data.
Python Implementation:
python
import pandas as pd
import numpy as np
import sweetviz as sv
from pandas_profiling import ProfileReport
import missingno as msno
def comprehensive_data_assessment(df, dataset_name="Dataset"):
"""
Perform comprehensive initial data assessment
"""
print(f"=== COMPREHENSIVE ASSESSMENT: {dataset_name} ===")
# Basic structure assessment
print(f"Shape: {df.shape}")
print(f"Memory usage: {df.memory_usage(deep=True).sum() / 1024**2:.2f} MB")
# Data types assessment
print("\n--- DATA TYPES ---")
dtype_summary = df.dtypes.value_counts()
for dtype, count in dtype_summary.items():
print(f"{dtype}: {count} columns")
# Missing values assessment
print("\n--- MISSING VALUES ---")
missing_stats = df.isnull().sum()
missing_percent = (missing_stats / len(df)) * 100
missing_summary = pd.DataFrame({
'Missing_Count': missing_stats,
'Missing_Percent': missing_percent
})
print(missing_summary[missing_summary['Missing_Count'] > 0])
# Statistical summary for numerical columns
print("\n--- NUMERICAL SUMMARY ---")
numerical_cols = df.select_dtypes(include=[np.number]).columns
if len(numerical_cols) > 0:
print(df[numerical_cols].describe())
# Categorical summary
print("\n--- CATEGORICAL SUMMARY ---")
categorical_cols = df.select_dtypes(include=['object']).columns
for col in categorical_cols:
print(f"\n{col}:")
print(f" Unique values: {df[col].nunique()}")
print(f" Most frequent: {df[col].mode().iloc[0] if not df[col].mode().empty else 'N/A'}")
return {
'shape': df.shape,
'memory_mb': df.memory_usage(deep=True).sum() / 1024**2,
'missing_summary': missing_summary,
'numerical_cols': numerical_cols.tolist(),
'categorical_cols': categorical_cols.tolist()
}
# Advanced visualization for data assessment
def visualize_data_assessment(df):
"""
Create comprehensive visual assessment
"""
import matplotlib.pyplot as plt
import seaborn as sns
fig, axes = plt.subplots(2, 2, figsize=(15, 12))
# Missing values matrix
msno.matrix(df, ax=axes[0,0])
axes[0,0].set_title('Missing Values Pattern')
# Data types distribution
dtype_counts = df.dtypes.value_counts()
axes[0,1].pie(dtype_counts.values, labels=dtype_counts.index, autopct='%1.1f%%')
axes[0,1].set_title('Data Types Distribution')
# Correlation heatmap for numerical data
numerical_df = df.select_dtypes(include=[np.number])
if len(numerical_df.columns) > 1:
sns.heatmap(numerical_df.corr(), annot=True, cmap='coolwarm', ax=axes[1,0])
axes[1,0].set_title('Numerical Features Correlation')
# Missing values by column
missing_by_col = df.isnull().sum()
axes[1,1].barh(missing_by_col.index, missing_by_col.values)
axes[1,1].set_title('Missing Values by Column')
plt.tight_layout()
plt.show()
# Example usage
df = pd.read_csv('your_dataset.csv')
assessment_results = comprehensive_data_assessment(df)
visualize_data_assessment(df)
# Generate automated profile report
profile = ProfileReport(df, title="Data Wrangling Assessment Report")
profile.to_file("data_assessment_report.html")R Implementation:
r
library(dplyr)
library(ggplot2)
library(DataExplorer)
library(summarytools)
library(visdat)
comprehensive_data_assessment <- function(df, dataset_name = "Dataset") {
cat("=== COMPREHENSIVE ASSESSMENT:", dataset_name, "===\n")
# Basic structure assessment
cat("Dimensions:", dim(df), "\n")
cat("Memory usage:", format(object.size(df), units = "MB"), "\n")
# Data types assessment
cat("\n--- DATA TYPES ---\n")
print(table(sapply(df, class)))
# Missing values assessment
cat("\n--- MISSING VALUES ---\n")
missing_summary <- data.frame(
Column = names(df),
Missing_Count = colSums(is.na(df)),
Missing_Percent = round(colSums(is.na(df)) / nrow(df) * 100, 2)
)
print(missing_summary[missing_summary$Missing_Count > 0, ])
# Statistical summary
cat("\n--- NUMERICAL SUMMARY ---\n")
numerical_cols <- df %>% select(where(is.numeric))
if (ncol(numerical_cols) > 0) {
print(summary(numerical_cols))
}
# Categorical summary
cat("\n--- CATEGORICAL SUMMARY ---\n")
categorical_cols <- df %>% select(where(is.character))
for (col in names(categorical_cols)) {
cat(col, ":\n")
cat(" Unique values:", n_distinct(df[[col]]), "\n")
cat(" Most frequent:", names(sort(table(df[[col]]), decreasing = TRUE))[1], "\n")
}
return(list(
dimensions = dim(df),
memory_usage = object.size(df),
missing_summary = missing_summary,
numerical_cols = names(numerical_cols),
categorical_cols = names(categorical_cols)
))
}
# Visual assessment in R
visualize_data_assessment <- function(df) {
# Missing values pattern
visdat::vis_miss(df)
# Data structure plot
DataExplorer::plot_intro(df)
# Correlation plot for numerical data
numerical_df <- df %>% select(where(is.numeric))
if (ncol(numerical_df) > 1) {
corr_matrix <- cor(numerical_df, use = "complete.obs")
corrplot::corrplot(corr_matrix, method = "color")
}
}
# Example usage
df <- read.csv("your_dataset.csv")
assessment_results <- comprehensive_data_assessment(df)
visualize_data_assessment(df)
# Generate comprehensive report
DataExplorer::create_report(df, output_file = "data_assessment_report.html")Step 2: Strategic Handling of Missing Values

Advanced Techniques for Missing Data Wrangling
Missing values represent one of the most common challenges in Data wrangling. How you handle missing data can significantly impact your analysis results. This step covers sophisticated approaches beyond simple deletion or mean imputation.
Python Implementation:
python
import pandas as pd
import numpy as np
from sklearn.impute import SimpleImputer, KNNImputer
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
import warnings
warnings.filterwarnings('ignore')
class AdvancedMissingValueHandler:
"""
Advanced missing value handling with multiple strategies
"""
def __init__(self):
self.imputation_models = {}
self.missing_patterns = {}
def analyze_missing_patterns(self, df):
"""
Analyze patterns and mechanisms of missing data
"""
self.missing_patterns = {
'missing_by_column': df.isnull().sum(),
'missing_by_row': df.isnull().sum(axis=1),
'missing_combinations': self._find_missing_combinations(df)
}
print("Missing Data Analysis:")
print(f"Total missing values: {df.isnull().sum().sum()}")
print(f"Percentage missing: {(df.isnull().sum().sum() / (df.shape[0] * df.shape[1])) * 100:.2f}%")
return self.missing_patterns
def _find_missing_combinations(self, df):
"""
Identify columns that tend to be missing together
"""
missing_corr = df.isnull().corr()
return missing_corr
def strategic_imputation(self, df, strategy='auto'):
"""
Implement strategic imputation based on data characteristics
"""
df_imputed = df.copy()
for column in df.columns:
missing_rate = df[column].isnull().mean()
# If missing rate is too high, consider dropping
if missing_rate > 0.8:
print(f"High missing rate in {column} ({missing_rate:.1%}) - consider dropping")
continue
# Choose imputation strategy based on data type and missing pattern
if df[column].dtype in ['float64', 'int64']:
if missing_rate < 0.05:
# For low missing rates, use mean/median
if strategy == 'median' or self._has_outliers(df[column]):
imputer = SimpleImputer(strategy='median')
else:
imputer = SimpleImputer(strategy='mean')
else:
# For higher missing rates, use more sophisticated methods
if strategy == 'knn':
imputer = KNNImputer(n_neighbors=5)
else:
imputer = IterativeImputer(max_iter=10, random_state=42)
# Fit and transform
df_imputed[column] = imputer.fit_transform(df[[column]]).ravel()
self.imputation_models[column] = imputer
elif df[column].dtype == 'object':
# For categorical data
if missing_rate < 0.1:
imputer = SimpleImputer(strategy='most_frequent')
else:
imputer = SimpleImputer(strategy='constant', fill_value='Unknown')
df_imputed[column] = imputer.fit_transform(df[[column]]).ravel()
self.imputation_models[column] = imputer
return df_imputed
def _has_outliers(self, series):
"""
Check if a series has significant outliers
"""
Q1 = series.quantile(0.25)
Q3 = series.quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
return ((series < lower_bound) | (series > upper_bound)).any()
def create_missing_indicators(self, df, threshold=0.05):
"""
Create indicator variables for missing patterns
"""
df_with_indicators = df.copy()
for column in df.columns:
missing_rate = df[column].isnull().mean()
if 0.05 <= missing_rate <= 0.8: # Meaningful missing pattern
indicator_name = f"{column}_missing"
df_with_indicators[indicator_name] = df[column].isnull().astype(int)
return df_with_indicators
# Example usage
handler = AdvancedMissingValueHandler()
missing_patterns = handler.analyze_missing_patterns(df)
# Create missing indicators for meaningful patterns
df_with_indicators = handler.create_missing_indicators(df)
# Perform strategic imputation
df_imputed = handler.strategic_imputation(df_with_indicators, strategy='auto')
print("Missing values before:", df.isnull().sum().sum())
print("Missing values after:", df_imputed.isnull().sum().sum())R Implementation:
r
library(mice)
library(VIM)
library(missForest)
library(dplyr)
advanced_missing_value_handler <- function(df) {
# Analyze missing patterns
missing_patterns <- list(
missing_by_column = colSums(is.na(df)),
missing_by_row = rowSums(is.na(df)),
missing_percentage = mean(is.na(df)) * 100
)
cat("Missing Data Analysis:\n")
cat("Total missing values:", sum(is.na(df)), "\n")
cat("Percentage missing:", mean(is.na(df)) * 100, "%\n")
# Visualize missing patterns
VIM::aggr(df, numbers = TRUE, sortVars = TRUE)
return(missing_patterns)
}
strategic_imputation <- function(df) {
df_imputed <- df
for (col in names(df)) {
missing_rate <- mean(is.na(df[[col]]))
if (missing_rate > 0.8) {
cat("High missing rate in", col, "(", round(missing_rate * 100, 1), "%) - consider dropping\n")
next
}
if (is.numeric(df[[col]])) {
if (missing_rate < 0.05) {
# Mean/median imputation for low missing rates
if (has_outliers(df[[col]])) {
df_imputed[[col]][is.na(df_imputed[[col]])] <- median(df[[col]], na.rm = TRUE)
} else {
df_imputed[[col]][is.na(df_imputed[[col]])] <- mean(df[[col]], na.rm = TRUE)
}
} else {
# Multiple imputation for higher rates
temp_df <- data.frame(df[[col]])
names(temp_df) <- col
imputed <- mice::mice(temp_df, method = "pmm", m = 1, printFlag = FALSE)
df_imputed[[col]] <- complete(imputed)[[col]]
}
} else if (is.character(df[[col]]) | is.factor(df[[col]])) {
# Categorical imputation
if (missing_rate < 0.1) {
mode_val <- names(sort(table(df[[col]]), decreasing = TRUE))[1]
df_imputed[[col]][is.na(df_imputed[[col]])] <- mode_val
} else {
df_imputed[[col]][is.na(df_imputed[[col]])] <- "Unknown"
}
}
}
return(df_imputed)
}
has_outliers <- function(x) {
if (!is.numeric(x)) return(FALSE)
Q1 <- quantile(x, 0.25, na.rm = TRUE)
Q3 <- quantile(x, 0.75, na.rm = TRUE)
IQR <- Q3 - Q1
lower_bound <- Q1 - 1.5 * IQR
upper_bound <- Q3 + 1.5 * IQR
return(any(x < lower_bound | x > upper_bound, na.rm = TRUE))
}
create_missing_indicators <- function(df, threshold = 0.05) {
df_with_indicators <- df
for (col in names(df)) {
missing_rate <- mean(is.na(df[[col]]))
if (missing_rate >= threshold & missing_rate <= 0.8) {
indicator_name <- paste0(col, "_missing")
df_with_indicators[[indicator_name]] <- as.integer(is.na(df[[col]]))
}
}
return(df_with_indicators)
}
# Example usage
missing_patterns <- advanced_missing_value_handler(df)
df_with_indicators <- create_missing_indicators(df)
df_imputed <- strategic_imputation(df_with_indicators)
cat("Missing values before:", sum(is.na(df)), "\n")
cat("Missing values after:", sum(is.na(df_imputed)), "\n")Step 3: Data Type Conversion and Validation

Ensuring Data Integrity Through Proper Typing
Correct data types are fundamental for accurate analysis and efficient computation. This step involves converting data to appropriate types and Data wrangling validating the conversions.
Python Implementation:
python
import pandas as pd
import numpy as np
from datetime import datetime
import re
class DataTypeValidator:
"""
Comprehensive data type conversion and validation
"""
def __init__(self):
self.conversion_log = []
self.validation_errors = []
def smart_type_conversion(self, df):
"""
Automatically detect and convert data types
"""
df_converted = df.copy()
for column in df.columns:
original_dtype = str(df[column].dtype)
# Skip if already optimal type
if self._is_optimal_type(df[column]):
continue
# Attempt different conversions
converted = False
# Try numeric conversion
if not converted:
numeric_converted = self._try_numeric_conversion(df[column])
if numeric_converted is not None:
df_converted[column] = numeric_converted
self.conversion_log.append({
'column': column,
'from': original_dtype,
'to': str(numeric_converted.dtype),
'method': 'numeric'
})
converted = True
# Try datetime conversion
if not converted and self._looks_like_date(df[column]):
datetime_converted = self._try_datetime_conversion(df[column])
if datetime_converted is not None:
df_converted[column] = datetime_converted
self.conversion_log.append({
'column': column,
'from': original_dtype,
'to': 'datetime64[ns]',
'method': 'datetime'
})
converted = True
# Try categorical conversion for low-cardinality strings
if not converted and df[column].dtype == 'object':
unique_ratio = df[column].nunique() / len(df[column])
if unique_ratio < 0.1: # Less than 10% unique values
df_converted[column] = df[column].astype('category')
self.conversion_log.append({
'column': column,
'from': original_dtype,
'to': 'category',
'method': 'categorical'
})
converted = True
return df_converted
def _is_optimal_type(self, series):
"""
Check if series already has optimal data type
"""
dtype = series.dtype
if dtype in [np.int64, np.float64, 'datetime64[ns]', 'category']:
return True
# For object type with low cardinality, category might be better
if dtype == 'object':
unique_ratio = series.nunique() / len(series)
if unique_ratio < 0.1:
return False
return False
def _try_numeric_conversion(self, series):
"""
Attempt to convert series to numeric type
"""
# First try direct conversion
try:
return pd.to_numeric(series, errors='raise')
except:
pass
# Try with cleaning
cleaned_series = series.astype(str).str.replace('[^\d.-]', '', regex=True)
try:
return pd.to_numeric(cleaned_series, errors='raise')
except:
return None
def _looks_like_date(self, series):
"""
Heuristic check if series contains date-like strings
"""
sample = series.dropna().head(100)
if len(sample) == 0:
return False
date_patterns = [
r'\d{1,2}/\d{1,2}/\d{2,4}',
r'\d{4}-\d{2}-\d{2}',
r'\d{1,2}-\d{1,2}-\d{2,4}'
]
date_like_count = 0
for value in sample.astype(str):
for pattern in date_patterns:
if re.match(pattern, value):
date_like_count += 1
break
return date_like_count / len(sample) > 0.5
def _try_datetime_conversion(self, series):
"""
Attempt to convert series to datetime
"""
try:
return pd.to_datetime(series, errors='raise')
except:
pass
# Try with different formats
for fmt in ['%Y-%m-%d', '%m/%d/%Y', '%d-%m-%Y', '%Y/%m/%d']:
try:
return pd.to_datetime(series, format=fmt, errors='coerce')
except:
continue
return None
def validate_data_integrity(self, df, rules):
"""
Validate data against business rules
"""
validation_results = {}
for column, rule in rules.items():
if column not in df.columns:
self.validation_errors.append(f"Column {column} not found")
continue
if 'min' in rule:
below_min = df[column] < rule['min']
if below_min.any():
validation_results[f"{column}_below_min"] = below_min.sum()
if 'max' in rule:
above_max = df[column] > rule['max']
if above_max.any():
validation_results[f"{column}_above_max"] = above_max.sum()
if 'allowed_values' in rule:
invalid_values = ~df[column].isin(rule['allowed_values'])
if invalid_values.any():
validation_results[f"{column}_invalid_values"] = invalid_values.sum()
return validation_results
# Example usage
validator = DataTypeValidator()
# Convert data types
df_typed = validator.smart_type_conversion(df)
print("Conversion log:")
for log in validator.conversion_log:
print(f" {log['column']}: {log['from']} -> {log['to']}")
# Define validation rules
validation_rules = {
'age': {'min': 0, 'max': 120},
'income': {'min': 0},
'category': {'allowed_values': ['A', 'B', 'C', 'D']}
}
# Validate data
validation_results = validator.validate_data_integrity(df_typed, validation_rules)
print("Validation results:", validation_results)R Implementation:
r
#Data wrangling
library(dplyr)
library(lubridate)
library(stringr)
smart_type_conversion <- function(df) {
df_converted <- df
conversion_log <- list()
for (col in names(df)) {
original_type <- class(df[[col]])
# Skip if already optimal type
if (is_optimal_type(df[[col]])) {
next
}
# Try numeric conversion
if (!is.numeric(df[[col]])) {
numeric_converted <- try_numeric_conversion(df[[col]])
if (!is.null(numeric_converted)) {
df_converted[[col]] <- numeric_converted
conversion_log[[col]] <- list(
from = original_type,
to = "numeric",
method = "numeric"
)
next
}
}
# Try date conversion
if (looks_like_date(df[[col]])) {
date_converted <- try_date_conversion(df[[col]])
if (!is.null(date_converted)) {
df_converted[[col]] <- date_converted
conversion_log[[col]] <- list(
from = original_type,
to = "Date",
method = "date"
)
next
}
}
# Convert to factor for low cardinality character columns
if (is.character(df[[col]])) {
unique_ratio <- n_distinct(df[[col]]) / length(df[[col]])
if (unique_ratio < 0.1) {
df_converted[[col]] <- as.factor(df[[col]])
conversion_log[[col]] <- list(
from = original_type,
to = "factor",
method = "categorical"
)
}
}
}
return(list(df = df_converted, log = conversion_log))
}
is_optimal_type <- function(x) {
if (is.numeric(x) | inherits(x, "Date") | is.factor(x)) {
return(TRUE)
}
if (is.character(x)) {
unique_ratio <- n_distinct(x) / length(x)
if (unique_ratio < 0.1) {
return(FALSE) # Should be factor
}
}
return(FALSE)
}
try_numeric_conversion <- function(x) {
# Try direct conversion
numeric_x <- suppressWarnings(as.numeric(x))
if (!any(is.na(numeric_x)) | sum(is.na(numeric_x)) == sum(is.na(x))) {
return(numeric_x)
}
# Try with cleaning
cleaned_x <- str_replace_all(as.character(x), "[^\\d.-]", "")
numeric_x <- suppressWarnings(as.numeric(cleaned_x))
if (!any(is.na(numeric_x)) | sum(is.na(numeric_x)) == sum(is.na(x))) {
return(numeric_x)
}
return(NULL)
}
looks_like_date <- function(x) {
sample_x <- na.omit(x)[1:min(100, length(na.omit(x)))]
if (length(sample_x) == 0) return(FALSE)
date_like_count <- 0
for (value in sample_x) {
if (grepl("\\d{1,2}/\\d{1,2}/\\d{2,4}", value) |
grepl("\\d{4}-\\d{2}-\\d{2}", value) |
grepl("\\d{1,2}-\\d{1,2}-\\d{2,4}", value)) {
date_like_count <- date_like_count + 1
}
}
return(date_like_count / length(sample_x) > 0.5)
}
try_date_conversion <- function(x) {
date_x <- suppressWarnings(as.Date(x))
if (!any(is.na(date_x)) | sum(is.na(date_x)) == sum(is.na(x))) {
return(date_x)
}
# Try different formats
formats <- c("%Y-%m-%d", "%m/%d/%Y", "%d-%m-%Y", "%Y/%m/%d")
for (fmt in formats) {
date_x <- suppressWarnings(as.Date(x, format = fmt))
if (!any(is.na(date_x)) | sum(is.na(date_x)) == sum(is.na(x))) {
return(date_x)
}
}
return(NULL)
}
validate_data_integrity <- function(df, rules) {
validation_results <- list()
for (col in names(rules)) {
if (!col %in% names(df)) {
warning(paste("Column", col, "not found"))
next
}
rule <- rules[[col]]
if (!is.null(rule$min)) {
below_min <- df[[col]] < rule$min
if (any(below_min, na.rm = TRUE)) {
validation_results[[paste0(col, "_below_min")]] <- sum(below_min, na.rm = TRUE)
}
}
if (!is.null(rule$max)) {
above_max <- df[[col]] > rule$max
if (any(above_max, na.rm = TRUE)) {
validation_results[[paste0(col, "_above_max")]] <- sum(above_max, na.rm = TRUE)
}
}
if (!is.null(rule$allowed_values)) {
invalid_values <- !df[[col]] %in% rule$allowed_values
if (any(invalid_values, na.rm = TRUE)) {
validation_results[[paste0(col, "_invalid_values")]] <- sum(invalid_values, na.rm = TRUE)
}
}
}
return(validation_results)
}
# Example usage
conversion_result <- smart_type_conversion(df)
df_typed <- conversion_result$df
print("Conversion log:")
print(conversion_result$log)
# Define validation rules
validation_rules <- list(
age = list(min = 0, max = 120),
income = list(min = 0),
category = list(allowed_values = c("A", "B", "C", "D"))
)
# Validate data
validation_results <- validate_data_integrity(df_typed, validation_rules)
print("Validation results:")
print(validation_results)
Step 4: Advanced Outlier Detection and Treatment
Sophisticated Approaches for Anomaly Detection
Outliers can significantly impact statistical analyses and machine learning models. This step covers advanced techniques for detecting and handling outliers in your Data wrangling pipeline.
Python Implementation:
python
import pandas as pd
import numpy as np
from sklearn.ensemble import IsolationForest
from sklearn.neighbors import LocalOutlierFactor
from sklearn.preprocessing import StandardScaler
import scipy.stats as stats
import matplotlib.pyplot as plt
import seaborn as sns
class AdvancedOutlierDetector:
"""
Comprehensive outlier detection using multiple methods
"""
def __init__(self):
self.detection_results = {}
self.scaler = StandardScaler()
def detect_outliers_comprehensive(self, df, numerical_columns=None):
"""
Detect outliers using multiple statistical methods
"""
if numerical_columns is None:
numerical_columns = df.select_dtypes(include=[np.number]).columns.tolist()
outlier_summary = {}
for column in numerical_columns:
column_data = df[column].dropna()
if len(column_data) < 10: # Skip columns with too few data points
continue
outliers = {
'zscore': self._zscore_outliers(column_data),
'iqr': self._iqr_outliers(column_data),
'isolation_forest': self._isolation_forest_outliers(column_data),
'lof': self._lof_outliers(column_data),
'modified_zscore': self._modified_zscore_outliers(column_data)
}
# Combine results from different methods
combined_outliers = self._combine_outlier_methods(outliers, column_data)
outlier_summary[column] = {
'methods': outliers,
'combined': combined_outliers,
'outlier_count': combined_outliers.sum(),
'outlier_percentage': (combined_outliers.sum() / len(column_data)) * 100
}
self.detection_results = outlier_summary
return outlier_summary
def _zscore_outliers(self, data, threshold=3):
"""
Detect outliers using Z-score method
"""
z_scores = np.abs(stats.zscore(data))
return z_scores > threshold
def _iqr_outliers(self, data):
"""
Detect outliers using Interquartile Range method
"""
Q1 = data.quantile(0.25)
Q3 = data.quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
return (data < lower_bound) | (data > upper_bound)
def _isolation_forest_outliers(self, data, contamination=0.1):
"""
Detect outliers using Isolation Forest
"""
if len(data) < 10:
return pd.Series([False] * len(data), index=data.index)
data_reshaped = data.values.reshape(-1, 1)
clf = IsolationForest(contamination=contamination, random_state=42)
outliers = clf.fit_predict(data_reshaped)
return outliers == -1
def _lof_outliers(self, data, contamination=0.1):
"""
Detect outliers using Local Outlier Factor
"""
if len(data) < 10:
return pd.Series([False] * len(data), index=data.index)
data_reshaped = data.values.reshape(-1, 1)
lof = LocalOutlierFactor(contamination=contamination)
outliers = lof.fit_predict(data_reshaped)
return outliers == -1
def _modified_zscore_outliers(self, data, threshold=3.5):
"""
Detect outliers using modified Z-score (more robust)
"""
median = np.median(data)
mad = np.median(np.abs(data - median))
if mad == 0:
mad = 1e-6 # Avoid division by zero
modified_z_scores = 0.6745 * (data - median) / mad
return np.abs(modified_z_scores) > threshold
def _combine_outlier_methods(self, outliers_dict, data):
"""
Combine results from multiple outlier detection methods
"""
# Create a voting system
methods = ['zscore', 'iqr', 'isolation_forest', 'lof', 'modified_zscore']
votes = pd.Series(0, index=data.index)
for method in methods:
if method in outliers_dict and outliers_dict[method] is not None:
votes += outliers_dict[method].astype(int)
# Consider outlier if detected by at least 2 methods
return votes >= 2
def treat_outliers(self, df, strategy='cap', numerical_columns=None):
"""
Treat outliers based on selected strategy
"""
if numerical_columns is None:
numerical_columns = df.select_dtypes(include=[np.number]).columns.tolist()
df_treated = df.copy()
for column in numerical_columns:
if column not in self.detection_results:
continue
column_data = df_treated[column].copy()
outliers = self.detection_results[column]['combined']
if strategy == 'remove':
df_treated = df_treated[~outliers]
elif strategy == 'cap':
# Cap outliers to IQR bounds
Q1 = column_data.quantile(0.25)
Q3 = column_data.quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
df_treated.loc[outliers & (column_data < lower_bound), column] = lower_bound
df_treated.loc[outliers & (column_data > upper_bound), column] = upper_bound
elif strategy == 'transform':
# Apply log transformation to reduce outlier impact
if (column_dataData wrangling is not merely a preliminary step in the data science workflow—it is the foundation upon which all successful data analysis and machine learning projects are built. Throughout this comprehensive guide, Data wrangling we’ve explored the ten essential steps that transform raw, messy data into clean, structured, and analysis-ready datasets. The journey from data assessment to final validation represents a critical investment that pays exponential dividends in the quality and reliability of your analytical outcomes.
The true power of effective Data wrangling lies in its ability to reveal hidden patterns, ensure data integrity, and create robust features that drive meaningful insights. By mastering these techniques in both Python and R, you equip yourself with the versatile toolkit needed to tackle diverse data challenges across different domains and project requirements. From handling missing values with sophisticated imputation strategies to detecting outliers using advanced statistical methods, each step in the Data wrangling process contributes to building a more accurate and trustworthy data foundation.
As the data landscape continues to evolve with increasing volume, variety, and velocity, the importance of systematic Data wrangling only grows more critical. The techniques covered in this guide—from automated data type conversion and validation to sophisticated feature engineering and data quality assessment—provide a scalable framework that adapts to datasets of any size and complexity. Remember that Data wrangling is both an art and a science: while the tools and techniques provide the methodology, domain knowledge and critical thinking guide their application.
Ultimately, the time invested in mastering Data wrangling is time saved in debugging models, explaining anomalous results, and rebuilding analyses. By implementing these ten essential steps consistently across your projects, you’ll not only produce more reliable results but also develop the disciplined approach Data wrangling that distinguishes professional data scientists. Let this guide serve as your comprehensive reference for transforming chaotic data into organized intelligence, enabling you to focus on what truly matters: extracting valuable insights and driving data-informed decisions.